Humans have been successful at treating a host of diseases. Yet some continue to elude medicine’s best attempts.
Tuberculosis killed 15 percent of the 9.6 million people who contracted it worldwide in 2014. Typhus has flared periodically throughout history, with deadly consequences. Rocky Mountain spotted fever is a tick-borne threat in North and South America. Pneumonia is common everywhere.
What unites these diseases is their ability to fend off antibiotics and continue to reproduce, through a mysterious molecular route that yields thymine, a DNA building block the bacteria need to survive and reproduce.
Here is a list of bacteria that use the gene thyX to replicate, and their associated diseases and conditions. Some also have a second pathway, using a gene found in humans—thyA—to reproduce.
Bacillus anthracis—anthrax
Borrelia burgdorferi—Lyme disease
Campylobacter jejuni—diarrhea
Chlamydia trachomatis—trachoma
Chlamydia pneumonia—pneumonia
Clostridium botulinum—botulism
Helicobacter pylori—stomach ulcer, gastric cancer
Mycobacterium leprae—leprosy
Mycobacterium tuberculosis—tuberculosis
Rickettsia prowazeki—typhus
Rickettsia rickettsii—Rocky Mountain spotted fever
Treponema pallidum—syphillis
Now, researchers at the University of Iowa have revealed how these diseases replicate by tracing the precise steps through which they use a gene absent in humans, called thyX, to code an enzyme to produce thymine. In a paper published online Jan. 28 in the journal Science, the Iowa chemists break down each stage in a rapid-fire chain of chemical reactions showing how thyX and the enzyme it encodes are used in the diseases’ DNA-production cycle.
The discovery could lead to the creation of non-toxic antibiotics that block the chemical reaction involving thyX, rather than relying on the current method of testing millions of drug compounds in the hopes of finding one that would faithfully kill each disease.
“We know a lot about these pathogens, but we didn’t know how the enzyme with them is catalyzing the reactions for DNA synthesis—the chemistry behind it,” says Amnon Kohen, a chemistry professor at the UI and corresponding author on the paper. “Now, we’re showing at the molecular level, the principal steps by which the thyX-encoded enzyme catalyzes the production of thymine’s precursor, thymidylate.”
ThyX has lurked, undetected, for eons. Scientists first spotted the gene when they noticed that thermophilic bacteria—ancient organisms that live at very high temperatures and pressures around deep-ocean vents—were able to produce thymine even though they didn't seem to have the genes to do it. In 2002, a research group in France pinpointed the heretofore mystery genes, and called them thyX. These new—or rather, ancient—genes seemed to produce thymine similar to the human gene thyA but had evolved separately.
No one knew why.

Kohen’s group dove deep into molecular chemistry to figure it out.
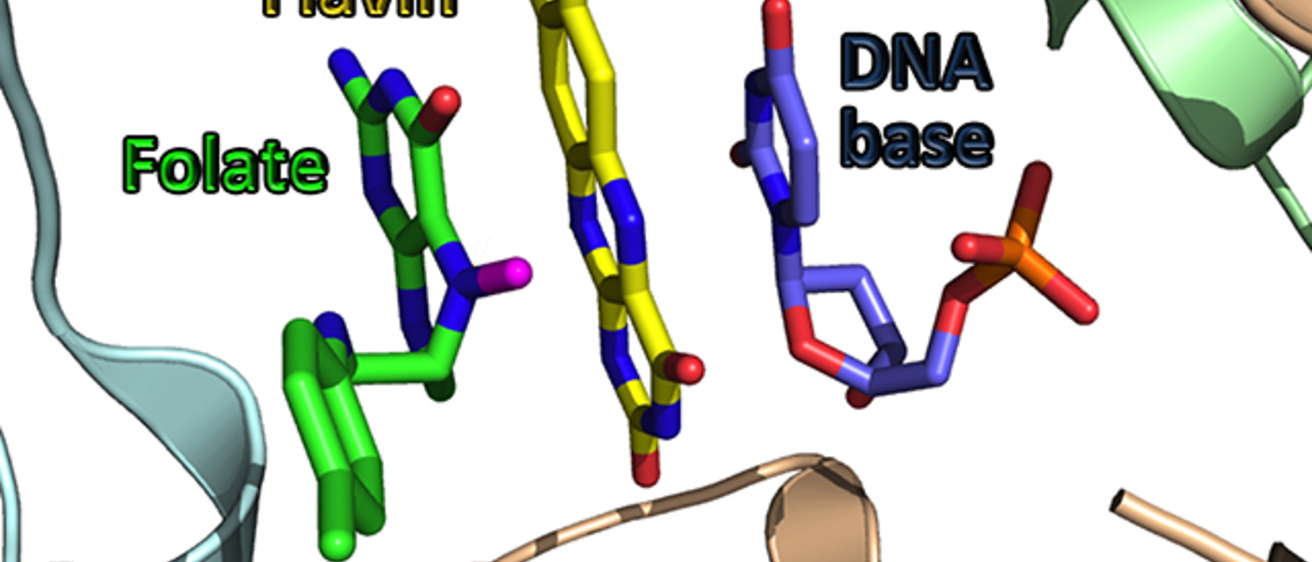
The team linked up with the UI’s Nuclear Magnetic Resonance facility to identify a critical intermediate of the reaction catalyzed by FDTS, an enzyme coded by thyX. When they compared that intermediate to those found in human enzymes, they found the paths to thymine were completely different. The human enzymes are encoded by folA and thyA genes, and their catalytic path involves a covalent bond activating one reactant, and direct chemistry between two reactants. The thyX-encoded FDTS, on the other hand, makes no bond with the reactant and conducts the chemistry through an enzymatic relay system (flavin).
“Actually, there are hardly any similarities between the classical mechanism found in humans and the newly discovered one,” Kohen says.
Several deadly diseases rely exclusively on thyX for survival and reproduction. Others, such as tuberculosis, can synthesize thymine with thyA or thyX, which makes them fiendishly difficult to eradicate because they can switch to another thymine pathway if one has been blocked. That explains why tuberculosis strains have become resistant to multiple drugs and thus difficult to contain.
Now that thyX’s role has been revealed, pharmaceutical companies can zero in on a product that gums up the cycle.
“Once it’s stuck, you’re dead in the water,” Kohen said.
Tatiana Mishanina, formerly with Kohen’s group and now at the University of Wisconsin–Madison, is the paper’s first author. Contributing authors are Liping Yu, director of the UI’s NMR facility, and members of Kohen’s group: Kalani Karunaratne, Dibyendu Mondal, John Corcoran, and Michael Choi.
The National Institutes of Health (grants R01 GM1 10775 and T32 GM008365) funded the study.